Research Overview
Our technological ability to both rapidly sequence and synthesize DNA has transformed the way we study biological systems. High-throughput technologies have led to large-scale connectivity maps of the biological gene networks in a field known as Systems Biology. A complimentary approach, Synthetic Biology, is emerging to synthetically build small networks of genes and then systematically increase complexity. These approaches lead to interesting and open questions including: What are the underlying design principles of a network of genes to perform a particular function? And how best can we rewire gene networks to build applications that shape environment, energy, and human health?
Our lab's research interests span a diverse array of topics that fall under three main areas:
Synthetic Biology
Our lab uses engineering principles to design and construct genetic circuits in microbes. In the past, we have engineered circuits to produce synchronized oscillations in a population of bacteria using quorum-sensing. We continue to work towards the development of genetic circuitry that control spatio-temporal behaviors in microbes.
Programming Bacteria for Cancer
The primary focus of the laboratory is to program bacteria as diagnostic and therapeutic agents for cancer. Using tools from synthetic biology, we engineer gene circuits that allow bacteria to communicate, reproduce, and express molecules inside of tumors. We use a methodology that spans from in silico mathematical modeling, to in vitro characterization, to in vivo animal testing in mice. Recently, our focus has been on bacteria therapeutics for breast cancer and its metastases, though we explore other microbiomes and human health applications.
Quantitative and Systems Biology
The design of new genetic circuits and microbial therapeutics are often inspired by native biological systems. Our lab utilizes mathematical modeling, next-generation sequencing, and microfluidic technologies to quantitatively understand the dynamics of gene networks and microbial populations in a variety of systems at a single cell level.
Introductory Videos
From selective breeding to genetic modification, our understanding of biology is now merging with the principles of engineering to bring us synthetic biology.
What if we could create a probiotic, edible bacteria that was "programmed" to find liver tumors? Tal Danino's insight exploits something we're just beginning to understand about bacteria: their power of quorum sensing, or doing something together once they reach critical mass. Danino, a TED Fellow, explains how quorum sensing works — and how clever bacteria working together could someday change cancer treatment.
By synchronising our clocks, we can coordinate our activities with people around the world. Now, scientists have engineered bacteria to synchronise their molecular timekeepers, creating the stunning fluorescent waves you see in this video.
Selected Abstracts
Synchronized cycles of bacterial lysis for in vivo delivery
M. Omar Din*, Tal Danino*, Arthur Prindle, Matthew Skalak, Jangir Selimkhanov, Kaitlin Allen, Ellixis Julio, Eta Atolia, Lev S. Tsimring, Sangeeta N. Bhatia, & Jeff Hasty. Nature 481, 39-44 (2016)
a, The circuit contains an activator and lysis plasmid. When the population reaches the quorum threshold at a critical AHL concentration, the luxI promoter drives the transcription of gene E for lysis,luxI, and sfGFP or luxCDABE as the reporter module. The luxI or the tac promoter also drives the transcription of the therapeutic gene for the stabilized circuit used in vivo. LuxR in this system is driven by the native luxR promoter. b, The main stages of each lysis cycle from seeding to quorum ‘firing’. Shown below the schematic depictions are typical time series images of the circuit-harbouring cells undergoing the three main stages of quorum firing in a microfluidic growth chamber. c, Fluorescence profile of a typical microfludic experiment. The estimated cell population trajectory reveals that lysis events correspond to peaks of sfGFP fluorescence. d, Period as a function of estimated flow velocity in the media channel of the microfluidic device and environmental temperature. Error bars indicate ± 1 s.d. for 13–50 peaks. These experiments were performed with strain 1, see Supplementary Information for complete strain information.
Summary: The widespread view of bacteria as strictly pathogenic has given way to an appreciation of the prevalence of some beneficial microbes within the human body. It is perhaps inevitable that some bacteria would evolve to preferentially grow in environments that harbour disease and thus provide a natural platform for the development of engineered therapies. Such therapies could benefit from bacteria that are programmed to limit bacterial growth while continually producing and releasing cytotoxic agents in situ. Here we engineer a clinically relevant bacterium to lyse synchronously at a threshold population density and to release genetically encoded cargo. Following quorum lysis, a small number of surviving bacteria reseed the growing population, thus leading to pulsatile delivery cycles. We used microfluidic devices to characterize the engineered lysis strain and we demonstrate its potential as a drug delivery platform via co-culture with human cancer cells in vitro. As a proof of principle, we tracked the bacterial population dynamics in ectopic syngeneic colorectal tumours in mice via a luminescent reporter. The lysis strain exhibits pulsatile population dynamics in vivo, with mean bacterial luminescence that remained two orders of magnitude lower than an unmodified strain. Finally, guided by previous findings that certain bacteria can enhance the efficacy of standard therapies, we orally administered the lysis strain alone or in combination with a clinical chemotherapeutic to a syngeneic mouse transplantation model of hepatic colorectal metastases. We found that the combination of both circuit-engineered bacteria and chemotherapy leads to a notable reduction of tumour activity along with a marked survival benefit over either therapy alone. Our approach establishes a methodology for leveraging the tools of synthetic biology to exploit the natural propensity for certain bacteria to colonize disease sites.
Programmable probiotics for detection of cancer in urine
Tal Danino*, Arthur Prindle*, Gabriel A. Kwong, Matthew Skalak, Howard Li, Kaitlin Allen, Jeff Hasty, and Sangeeta Bhatia. "Programmable probiotics for detection of cancer in urine." Science Translational Medicine Vol. 7, Issue 289, p. 289ra84.
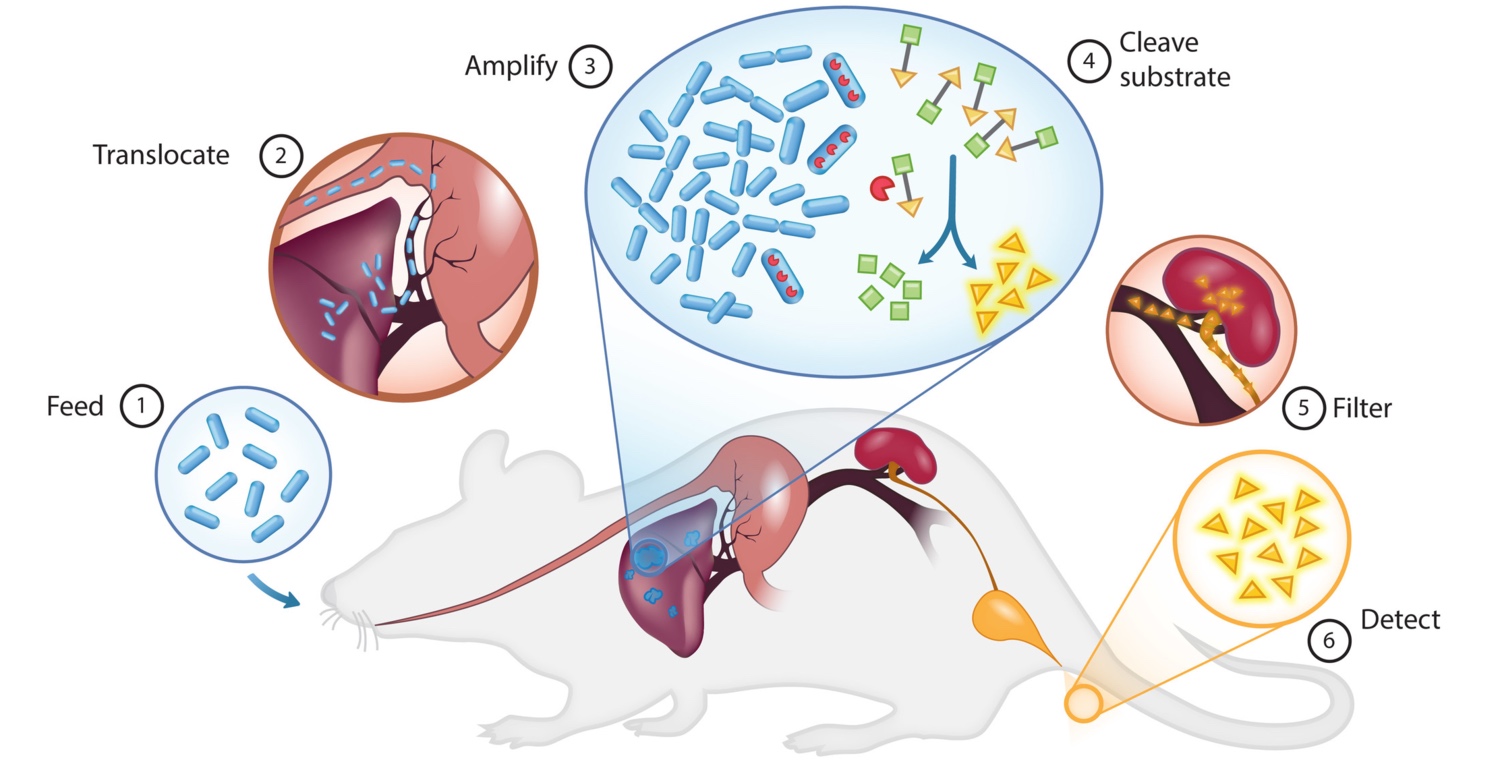
PROP-Z (Programmable Probiotics with lacZ) for noninvasive cancer detection. The PROP-Z diagnostic platform is made up of probiotic EcN bacteria transformed with a dual-stabilized, high-expression lacZ vector as well as a genomically integrated luxCDABE cassette that allows for luminescent visualization without providing exogenous luciferin (blue). (1) PROP-Z is delivered orally. (2) Probiotics rapidly (within 24 hours) translocate across the gastrointestinal tract and (3) specifically amplify within metastatic tumors present in the liver. (4) PROP-Z expresses high levels of the enzyme lacZ (red), which can cleave systemically injected, cleavable substrates (green and yellow). Cleavage products of the substrates (yellow) filter through the renal system (5) into the urine for detection (6).
Summary: Rapid advances in the forward engineering of genetic circuitry in living cells has positioned synthetic biology as a potential means to solve numerous biomedical problems, including disease diagnosis and therapy. One challenge in exploiting synthetic biology for translational applications is to engineer microbes that are well tolerated by patients and seamlessly integrate with existing clinical methods. We use the safe and widely used probiotic Escherichia coli Nissle 1917 to develop an orally administered diagnostic that can noninvasively indicate the presence of liver metastasis by producing easily detectable signals in urine. Our microbial diagnostic generated a high-contrast urine signal through selective expansion in liver metastases (106-fold enrichment) and high expression of a lacZ reporter maintained by engineering a stable plasmid system. The lacZ reporter cleaves a substrate to produce a small molecule that can be detected in urine. E. coli Nissle 1917 robustly colonized tumor tissue in rodent models of liver metastasis after oral delivery but did not colonize healthy organs or fibrotic liver tissue. We saw no deleterious health effects on the mice for more than 12 months after oral delivery. Our results demonstrate that probiotics can be programmed to safely and selectively deliver synthetic gene circuits to diseased tissue microenvironments in vivo.
A synchronized quorum of genetic clocks
Tal Danino*, Octavio Mondragón-Palomino*, Lev Tsimring, and Jeff Hasty. "A synchronized quorum of genetic clocks." Nature 463, no. 7279 (2010): 326-330.
Synchronized genetic clocks. (A) Network diagram. The luxI promoter drives production of the luxI, aiiA and yemGFP genes in three identical transcriptional modules. LuxI enzymatically produces a small molecule AHL, which can diffuse outside of the cell membrane and into neighbouring cells, activating the luxI promoter. AiiA negatively regulates the circuit by acting as an effective protease for AHL. (B) The "Supernova". Fluorescence microscopy overlay of a growing colony in a microfluidic device at the time it reaches a quorum. (C) Fluorescence slices of a typical experimental run demonstrate synchronization of oscillations in a population of E. coli residing in the microfluidic device (Movie in Videos page). Inset in the first snapshot is a ×100 magnification of cells.
Summary: The engineering of genetic circuits with predictive functionality in living cells represents a defining focus of the expanding field of synthetic biology. This focus was elegantly set in motion a decade ago with the design and construction of a genetic toggle switch and an oscillator, with subsequent highlights that have included circuits capable of pattern generation, noise shaping, edge detection and event counting. Here we describe an engineered gene network with global intercellular coupling that is capable of generating synchronized oscillations in a growing population of cells. Using microfluidic devices tailored for cellular populations at differing length scales, we investigate the collective synchronization properties along with spatiotemporal waves occurring at millimetre scales. We use computational modelling to describe quantitatively the observed dependence of the period and amplitude of the bulk oscillations on the flow rate. The synchronized genetic clock sets the stage for the use of microbes in the creation of a macroscopic biosensor with an oscillatory output. Furthermore, it provides a specific model system for the generation of a mechanistic description of emergent coordinated behaviour at the colony level.
Entrainment of a population of synthetic genetic oscillators
Octavio Mondragón-Palomino, Tal Danino, Jangir Selimkhanov, Lev Tsimring, and Jeff Hasty. "Entrainment of a population of synthetic genetic oscillators." Science 333, no. 6047 (2011): 1315-1319.
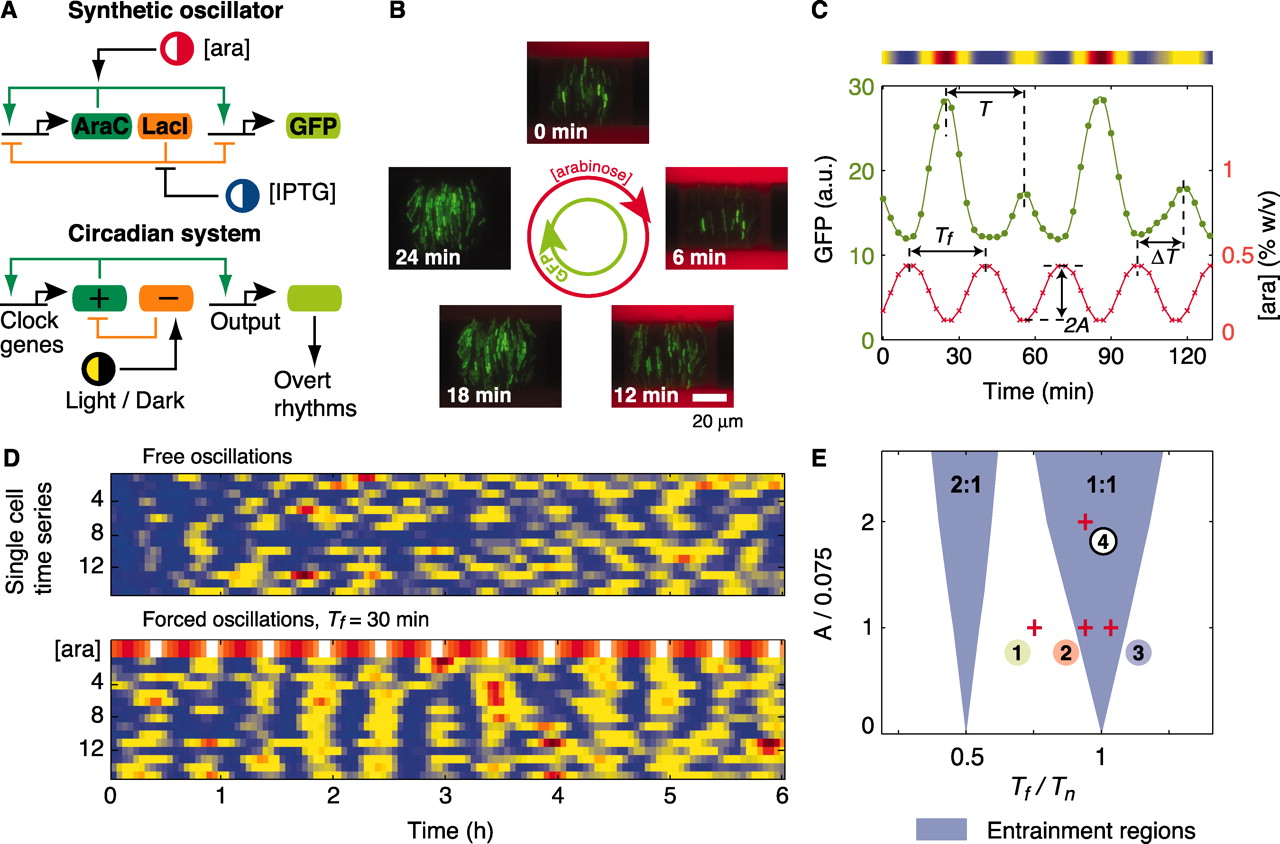
Entrainment of genetic oscillators. (A) Architectures of eukaryotic circadian clocks and bacterial synthetic oscillators contain positive- and negative-feedback loops that are sensitive to external stimuli. (B) Fluorescence images from a time-lapse experiment show coherent GFP oscillations (green) in a colony of single-cell oscillators subject to a 30-min cycle of arabinose (red) (movie S1). (C)Fluorescence time series of a single-cell oscillator (green). The concentration of arabinose (red) changes sinusoidally according to [ara](t) = 0.3 + Asin(2πt/Tf) [percent weight/volume (% w/v)], with A = 0.15% and Tf = 30 min. The intensity plot above the graph corresponds to the cell trace. a.u., arbitrary units. (D) Fluorescence intensity plots of free-running and forced oscillators. (E) Entrainment regions indicate which forcing periods (Tf) and amplitudes (A) result in locking of the oscillator according to a deterministic model (SOM text). Entrainment of order 2:1 means that two oscillation peaks are observed for one peak of arabinose.
Summary: Biological clocks are self-sustained oscillators that adjust their phase to the daily environmental cycles in a process known as entrainment. Molecular dissection and mathematical modeling of biological oscillators have progressed quite far, but quantitative insights on the entrainment of clocks are relatively sparse. We simultaneously tracked the phases of hundreds of synthetic genetic oscillators relative to a common external stimulus to map the entrainment regions predicted by a detailed model of the clock. Synthetic oscillators were frequency-locked in wide intervals of the external period and showed higher-order resonance. Computational simulations indicated that natural oscillators may contain a positive-feedback loop to robustly adapt to environmental cycles.
A sensing array of radically coupled genetic "biopixels."
Arthur Prindle, Phillip Samayoa, Ivan Razinkov, Tal Danino, Lev S. Tsimring, and Jeff Hasty. "A sensing array of radically coupled genetic "biopixels." Nature 481, no. 7379 (2012): 39-44.
Bacteria "biopixels". (A) Network diagram. The luxI promoter drives expression of luxI, aiiA, ndh and sfGFP (superfolder variant of GFP) in four identical transcription modules. The quorum-sensing genes luxI and aiiA generate synchronized oscillations within a colony via AHL. The ndh gene codes for NDH-2, an enzyme that generates H2O2 vapour, which is an additional activator of the luxI promoter. H2O2 is capable of migrating between colonies and synchronizing them. (B) Conceptual design of the sensing array. AHL diffuses within colonies while H2O2 migrates between adjacent colonies through the PDMS. Arsenite-containing media is passed in through the parallel feeding channels. (C) Fluorescent image of an array of 500 E. coli biopixels containing about 2.5 million cells. Inset, bright-field and fluorescent images display a biopixel of 5,000 cells. (D) Heat map and trajectories depicting time-lapse output of 500 individual biopixels undergoing rapid synchronization. Sampling time is 2 min.
Summary: Although there has been considerable progress in the development of engineering principles for synthetic biology, a substantial challenge is the construction of robust circuits in a noisy cellular environment. Such an environment leads to considerable intercellular variability in circuit behaviour, which can hinder functionality at the colony level. Here we engineer the synchronization of thousands of oscillating colony ‘biopixels’ over centimetre-length scales through the use of synergistic intercellular coupling involving quorum sensing within a colony and gas-phase redox signalling between colonies. We use this platform to construct a liquid crystal display (LCD)-like macroscopic clock that can be used to sense arsenic via modulation of the oscillatory period. Given the repertoire of sensing capabilities of bacteria such as Escherichia coli, the ability to coordinate their behaviour over large length scales sets the stage for the construction of low cost genetic biosensors that are capable of detecting heavy metals and pathogens in the field.
[PDF Link]
In vivo gene expression dynamics of tumor-targeted bacteria
Tal Danino, Justin Lo, Arthur Prindle, Jeff Hasty, and Sangeeta N. Bhatia. "In vivo gene expression dynamics of tumor-targeted bacteria." ACS Synthetic Biology 1, no. 10 (2012): 465-470.
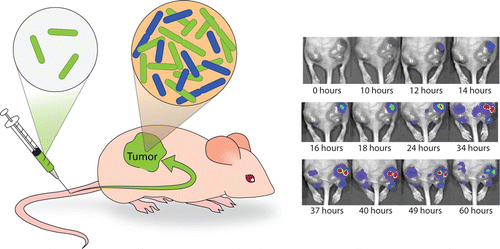
Dynamics of Tumor-Targeted Bacteria. (Left) S. typhimurium are injected via tail vein into nude mice and localize to subcutaneous tumors where they replicate. (Right) Sequence of IVIS images for an S.Typhimurium strain with a luminescent plasmid at 10^6 dosage over the course of 60 h. The IVIS signal rises due to rapid bacterial growth and then decays due to plasmid loss and luciferase instability.
Summary: The engineering of bacteria to controllably deliver therapeutics is an attractive application for synthetic biology. While most synthetic gene networks have been explored within microbes, there is a need for further characterization of in vivo circuit behavior in the context of applications where the host microbes are actively being investigated for efficacy and safety, such as tumor drug delivery. One major hurdle is that culture-based selective pressures are absent in vivo, leading to strain-dependent instability of plasmid-based networks over time. Here, we experimentally characterize the dynamics of in vivo plasmid instability using attenuated strains of S. typhimurium and real-time monitoring of luminescent reporters. Computational modeling described the effects of growth rate and dosage on live-imaging signals generated by internal bacterial populations. This understanding will allow us to harness the transient nature of plasmid-based networks to create tunable temporal release profiles that reduce dosage requirements and increase the safety of bacterial therapies.
[PDF Link]
In-silico patterning of vascular mesenchymal cells in three dimensions
Tal Danino, Dmitri Volfson, Sangeeta N. Bhatia, Lev Tsimring, and Jeff Hasty. "In-silico patterning of vascular mesenchymal cells in three dimensions." Plos one 6, no. 5 (2011): e20182
3d pattern formation of vascular mesenchymal cells. (A) Interactions between BMP-2, MGP, and cells in culture. The binding of a BMP-2 dimer to receptors R and S stimulates production of BMP-2 and MGP, while the inding of MGP to BMP-2 outside of the cell prevents this process. (B) The derived model shows spherical spots (k = 0.2,c = 0.12), tubes (k = 0.2,c = 0.04), and sheet-like structures (k = 0.8,c = 0.04) by varying k and c. The parameters used were D = 0.005, q = 0.003, K = 0.25, B = 1.1, γ = 600 and the box length of the simulation is equivalent to 1 cm. The lowest values were made transparent for clarity while red color indicates higher cell densitywhile blue indicates low.
Summary: Cells organize in complex three-dimensional patterns by interacting with proteins along with the surrounding extracellular matrix. This organization provides the mechanical and chemical cues that ultimately influence a cell's differentiation and function. Here, we computationally investigate the pattern formation process of vascular mesenchymal cells arising from their interaction with Bone Morphogenic Protein-2 (BMP-2) and its inhibitor, Matrix Gla Protein (MGP). Using a first-principles approach, we derive a reaction-diffusion model based on the biochemical interactions of BMP-2, MGP and cells. Simulations of the model exhibit a wide variety of three-dimensional patterns not observed in a two-dimensional analysis. We demonstrate the emergence of three types of patterns: spheres, tubes, and sheets, and show that the patterns can be tuned by modifying parameters in the model such as the degradation rates of proteins and chemotactic coefficient of cells. Our model may be useful for improved engineering of three-dimensional tissue structures as well as for understanding three dimensional microenvironments in developmental processes.